Large Scale Computation with Pegasus
Objectives
- Learn how to use Pegasus to complete a set of computational tasks.
Overview
Pegasus can handle millions of computational tasks and takes care of managing input/output files for you. In this section, we will get an overview of Pegasus with an example workflow, which calculates the word frequencies of a set of inputs. This demonstrates how to set up a workflow to run a given executable for each input in a given directory.
Introduction
Pegasus enables scientists to construct workflows in abstract terms without worrying about the details of the underlying execution environment or the particulars of the low-level specifications required by the middleware. Some of the advantages of using Pegasus includes:
-
Performance - The Pegasus mapper can reorder, group, and prioritize tasks in order to increase the overall workflow performance.
-
Scalability - Pegasus can easily scale both the size of the workflow, and the resources that the workflow is distributed over. Pegasus runs workflows ranging from just a few computational tasks up to 1 million. The number of resources involved in executing a workflow can scale as needed without any impediments to performance.
-
Portability / Reuse - User created workflows can easily be run in different environments without alteration. Pegasus currently runs workflows on top of Condor, Grid infrastrucutures such as Open Science Grid and TeraGrid, Amazon EC2, Nimbus, and many campus clusters. The same workflow can run on a single system or across a heterogeneous set of resources.
-
Data Management - Pegasus handles replica selection, data transfers and output registrations in data catalogs. These tasks are added to a workflow as auxiliary jobs by the Pegasus planner.
-
Reliability - Jobs and data transfers are automatically retried in case of failures. Debugging tools such as pegasus-analyzer helps the user to debug the workflow in case of non-recoverable failures.
-
Provenance - By default, all jobs in Pegasus are launched via the kickstart process that captures runtime provenance of the job and helps in debugging. The provenance data is collected in a database, and the data can be summaries with tools such as pegasus-statistics, pegasus-plots, or directly with SQL queries.
-
Error Recovery - When errors occur, Pegasus tries to recover when possible by retrying tasks, by retrying the entire workflow, by providing workflow-level checkpointing, by re-mapping portions of the workflow, by trying alternative data sources for staging data, and, when all else fails, by providing a rescue workflow containing a description of only the work that remains to be done.
As mentioned earlier, OSG has no read/write enabled shared file system across the resources. Jobs are required to either bring inputs along with the job, or as part of the job stage the inputs from a remote location. The following examples highlight how Pegasus can be used to manage workloads in such an environment by providing an abstraction layer around things like data movements and job retries, enabling the users to run larger workloads, spending less time developing job management tool and babysitting their computations.
Pegasus workflows have 4 components:
-
DAX - Abstract workflow description containing compute steps and dependencies between the steps. This is called abstract because it does not contain data locations and available software. The DAX format is XML, but it is most commonly generated via the provided APIS (documentation). Python, Java and Perl APIs are available.
-
Transformation Catalog - Specifies locations of software used by the workflow
-
Replica Catalog - Specifies locations of input data
-
Site Catalog - Describes the execution environment
However, for simple workflows, the transformation and replica catalog can be contained inside the DAX, and to further simplify the setup, the following examples generate the site catalog on the fly. This means that the user really only has to be concerned about creating the DAX. For details, please refer to the Pegasus documentation
The DAX and the catalogs are used as inputs to the Pegasus planning process. This takes the abstract workflow, and plans it into an executable workflow. The following picture illustrates how a two node abstract workflow could be planned for a particular execution environment:
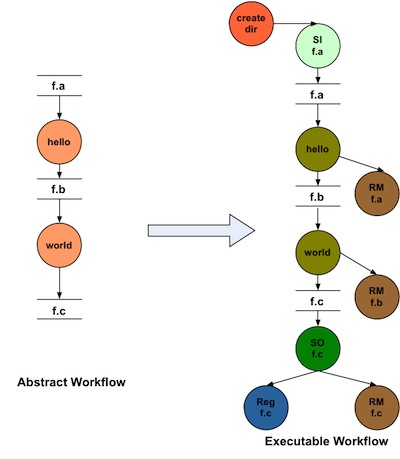
wordfreq example
The necessary files are available to the user by invoking the tutorial command.
$ source osg_oasis_init
$ module load pegasus/4.6.0dev
$ cd $WORK
$ tutorial pegasus
$ cd tutorial-pegasus/wordfreq-workflow
In the wordfreq-workflow directory, you will find:
- inputs/ (directory)
- dax-generator.py
- pegasusrc
- submit
- wordfreq
The inputs/ directory contains 6 public domain ebooks. The application in this example is wordfreq, which takes in a text file, does a word frequency analysis, and outputs a file with the frequency table. The task of the workflow is to run wordfreq on each book in the inputs/ directory. A set of independent tasks like this is a common use case on OSG and this workflow can be thought of as template for such problems. For example, instead of wordfreq on ebooks, the application could be protein folding on a set of input structures.
When invoked, the DAX generator (dax-generator.py) loops over the inputs/ directory and creates compute steps for each input. As the wordfreq application only has simple inputs/outputs, and no job dependencies, the DAX generator is very simple. See the dax-generator.py file below:
#!/usr/bin/env python
from Pegasus.DAX3 import *
import sys
import os
base_dir = os.getcwd()
# Create a abstract dag
dax = ADAG("wordfreq-workflow")
# Add executables to the DAX-level replica catalog
wordfreq = Executable(name="wordfreq", arch="x86_64", installed=False)
wordfreq.addPFN(PFN("file://" + base_dir + "/wordfreq", "local"))
wordfreq.addProfile(Profile(Namespace.PEGASUS, "clusters.size", 1))
dax.addExecutable(wordfreq)
# add jobs, one for each input file
inputs_dir = base_dir + "/inputs"
for in_name in os.listdir(inputs_dir):
# Add input file to the DAX-level replica catalog
in_file = File(in_name)
in_file.addPFN(PFN("file://" + inputs_dir + "/" + in_name, "local"))
dax.addFile(in_file)
# Add job
wordfreq_job = Job(name="wordfreq")
out_file = File(in_name + ".out")
wordfreq_job.addArguments(in_file, out_file)
wordfreq_job.uses(in_file, link=Link.INPUT)
wordfreq_job.uses(out_file, link=Link.OUTPUT)
dax.addJob(wordfreq_job)
# Write the DAX to stdout
f = open("dax.xml", "w")
dax.writeXML(f)
f.close()
Note how the DAX is devoid of data movement and job details. These are added by Pegasus when the DAX is planned to an executable workflow, and provides the higher level abstraction mentioned earlier.
In the tarball there is also a submit script. This is a convenience script written in bash, and it performs three steps: runs the DAX generator, generates a site catalog, and plans/submits the workflow for execution. The site catalog does not really have to be created every time we plan/submit a workflow, but in this case we have a workflow which is used by different users, so changing the paths to scratch and output filesystems on the fly makes the workflow easier to share.
Submitting
A new workflow can be submitted using the provided submit script:
$ ./submit
2016.01.05 13:19:02.128 CST:
2016.01.05 13:19:02.133 CST: -----------------------------------------------------------------------
2016.01.05 13:19:02.139 CST: File for submitting this DAG to Condor : wordfreq-workflow-0.dag.condor.sub
2016.01.05 13:19:02.144 CST: Log of DAGMan debugging messages : wordfreq-workflow-0.dag.dagman.out
2016.01.05 13:19:02.149 CST: Log of Condor library output : wordfreq-workflow-0.dag.lib.out
2016.01.05 13:19:02.155 CST: Log of Condor library error messages : wordfreq-workflow-0.dag.lib.err
2016.01.05 13:19:02.160 CST: Log of the life of condor_dagman itself : wordfreq-workflow-0.dag.dagman.log
2016.01.05 13:19:02.165 CST:
2016.01.05 13:19:02.181 CST: -----------------------------------------------------------------------
2016.01.05 13:19:03.458 CST: Your database is compatible with Pegasus version: 4.6.0
2016.01.05 13:19:03.588 CST: Submitting to condor wordfreq-workflow-0.dag.condor.sub
2016.01.05 13:19:03.755 CST: Submitting job(s).
2016.01.05 13:19:03.761 CST: 1 job(s) submitted to cluster 76355.
2016.01.05 13:19:03.766 CST:
2016.01.05 13:19:03.771 CST: Your workflow has been started and is running in the base directory:
2016.01.05 13:19:03.776 CST:
2016.01.05 13:19:03.782 CST: /work/bockelman/rynge/workflows/runs/1452021527
2016.01.05 13:19:03.787 CST:
2016.01.05 13:19:03.792 CST: *** To monitor the workflow you can run ***
2016.01.05 13:19:03.797 CST:
2016.01.05 13:19:03.802 CST: pegasus-status -l /work/bockelman/rynge/workflows/runs/1452021527
2016.01.05 13:19:03.808 CST:
2016.01.05 13:19:03.813 CST: *** To remove your workflow run ***
2016.01.05 13:19:03.818 CST:
2016.01.05 13:19:03.823 CST: pegasus-remove /work/bockelman/rynge/workflows/runs/1452021527
2016.01.05 13:19:03.829 CST:
2016.01.05 13:19:04.012 CST: Time taken to execute is 15.74 seconds
Note that Pegasus uses a directory as the handle to a workflow. Whereas a job id is only valid while the job is in the queue, a directory is a a valid handle before, during and after workflow execution.
Monitoring / Debugging
Use the directory provided in the output of the previous command to check on the status of your workflow:
$ pegasus-status [wfdir]
STAT IN_STATE JOB
Run 00:53 wordfreq-workflow-0
Idle 00:08 ┣━wordfreq_ID0000001
Idle 00:08 ┣━wordfreq_ID0000005
Idle 00:03 ┣━wordfreq_ID0000003
Idle 00:02 ┣━wordfreq_ID0000002
Idle 00:02 ┣━wordfreq_ID0000006
Idle 00:02 ┗━wordfreq_ID0000004
Summary: 7 Condor jobs total (I:6 R:1)
UNRDY READY PRE IN_Q POST DONE FAIL %DONE STATE DAGNAME
7 0 0 6 0 4 0 23.5 Running *wordfreq-workflow-0.dag
Summary: 1 DAG total (Running:1)
While not covered in this tutorial, if the workflow fails, pegasus-analyzer can be run to get a summary of what failed. This is usually more helpful than trying to look at logs and outputs of tasks.
Once the workflow is done, you can find outputs under $WORK/workflows/outputs/
Statistics
Once the workflow has completed (see the output of pegasus-status), you can find runtime statistics
using the pegasus-statistics command. Again, use the submit time provided directory as argument:
$ pegasus-statistics [wfdir]
#
# Pegasus Workflow Management System - http://pegasus.isi.edu
#
# Workflow summary:
# Summary of the workflow execution. It shows total
# tasks/jobs/sub workflows run, how many succeeded/failed etc.
# In case of hierarchical workflow the calculation shows the
# statistics across all the sub workflows.It shows the following
# statistics about tasks, jobs and sub workflows.
# * Succeeded - total count of succeeded tasks/jobs/sub workflows.
# * Failed - total count of failed tasks/jobs/sub workflows.
# * Incomplete - total count of tasks/jobs/sub workflows that are
# not in succeeded or failed state. This includes all the jobs
# that are not submitted, submitted but not completed etc. This
# is calculated as difference between 'total' count and sum of
# 'succeeded' and 'failed' count.
# * Total - total count of tasks/jobs/sub workflows.
# * Retries - total retry count of tasks/jobs/sub workflows.
# * Total+Retries - total count of tasks/jobs/sub workflows executed
# during workflow run. This is the cumulative of retries,
# succeeded and failed count.
# Workflow wall time:
# The wall time from the start of the workflow execution to the end as
# reported by the DAGMAN.In case of rescue dag the value is the
# cumulative of all retries.
# Cumulative job wall time:
# The sum of the wall time of all jobs as reported by kickstart.
# In case of job retries the value is the cumulative of all retries.
# For workflows having sub workflow jobs (i.e SUBDAG and SUBDAX jobs),
# the wall time value includes jobs from the sub workflows as well.
# Cumulative job wall time as seen from submit side:
# The sum of the wall time of all jobs as reported by DAGMan.
# This is similar to the regular cumulative job wall time, but includes
# job management overhead and delays. In case of job retries the value
# is the cumulative of all retries. For workflows having sub workflow
# jobs (i.e SUBDAG and SUBDAX jobs), the wall time value includes jobs
# from the sub workflows as well.
# Cumulative job badput wall time:
# The sum of the wall time of all failed jobs as reported by kickstart.
# In case of job retries the value is the cumulative of all retries.
# For workflows having sub workflow jobs (i.e SUBDAG and SUBDAX jobs),
# the wall time value includes jobs from the sub workflows as well.
# Cumulative job badput wall time as seen from submit side:
# The sum of the wall time of all failed jobs as reported by DAGMan.
# This is similar to the regular cumulative job badput wall time, but includes
# job management overhead and delays. In case of job retries the value
# is the cumulative of all retries. For workflows having sub workflow
# jobs (i.e SUBDAG and SUBDAX jobs), the wall time value includes jobs
# from the sub workflows as well.
------------------------------------------------------------------------------
Type Succeeded Failed Incomplete Total Retries Total+Retries
Tasks 6 0 0 6 0 6
Jobs 17 0 0 17 0 17
Sub-Workflows 0 0 0 0 0 0
------------------------------------------------------------------------------
Workflow wall time : 2 mins, 23 secs
Cumulative job wall time : 48 secs
Cumulative job wall time as seen from submit side : 1 min, 13 secs
Cumulative job badput wall time :
Cumulative job badput wall time as seen from submit side :
For real life computations, we expect the cumulative job wall time to be much greater thatn the workflow wall time. For example:
Workflow wall time : 18 hours, 32 mins
Cumulative job wall time : 346 years, 23 days
On your own
- Start a new workflow with a larger input data set. Copy all the files in the
many-more-inputsdirectory to theinputsdirectory (cp many-more-inputs/* inputs/), and run./submit. Check the status withpegasus-status- A nice feature is that failed/stopped workflows can be recovered from where they left of. While the workflow is running, use
pegasus-remove [dir]to stop the workflow. Wait for the jobs to leave the queue, and then usepegasus-run [dir]to start the workflow up again.
Keypoints
-
Pegasus requires dax.xml, sites.xml and pegasus.conf files. These files contain the information about executable, input and output files and the relation between them while executing the jobs.
-
It is convenient to generate the xml files via scripts. In our example, dax.xml is generated via python script and sites.xml is generated via bash script.
-
To implement a new workflow, edit the existing dax-generator, sites-generator and submit scripts.
