Handling Job Dependencies - DAGMan
Objectives
- Learn about Workflow Management
- Learn how to use DAGMan to complete a set of computational jobs
Overview
In scientific computing, one may have to perform several computational tasks or data manipulations that are inter dependent. Workflow management systems help to deal with such tasks or data manipulations. DAGMan is a workflow management system developed for distributed high throughput computing. DAGMan (Directed Acyclic Graph Manager) handles computational jobs that are mapped as a directed acyclic graph. Cyclic graph forms loop while acyclic graph does not form loop. Directed acyclic graph does not form loop and the nodes (jobs) are connected along specific direction. In this section, we will learn how to apply DAGMan to run a set of molecular dynamics (MD) simulations.
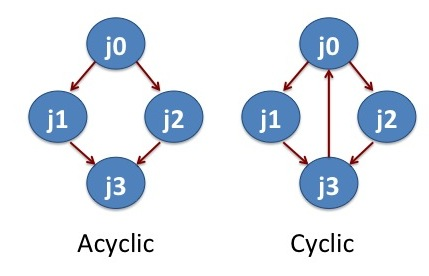
Running MD Simulation with DAGMan
At present, the recommended execution time to run a condor job on OSG is about 2-3 hours. Jobs requiring more than 2-3 hours, need to be submitted with the restart files. Manually submitting small jobs repeatedly with restart files may not be practical in many situations. DAGMan offers an elegant and simple solution to run the set of jobs. With the DAGMan script one could run a long time scale MD simulations of biomolecules.
Linear DAG
In our first example, we will break the MD simulation in four steps and run it through the DAGMan script. NAMD software is used to run each MD simulation. For the sake of simplicity, the MD simulations run only for few integration steps to consume less computational time but demonstrate the ability of DAGMan.
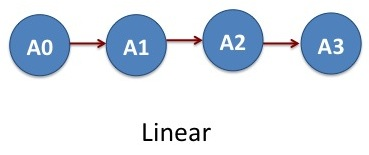
Say we have created four MD jobs: A0, A1, A2 and A3 that we want to run one after another and combine the results. This means that the output files from the job A0 serves as an input for the job A1 and so forth. The input and output dependencies of the jobs are such that they need to be progressed in a linear fashion: A0–>A1–>A2–>A3. These set of jobs clearly represents an acyclic graph. In DAGMan language, job A0 is parent of job A1, job A1 is parent of A2 and job A3 is parent of A4.
First login and get the DAGMan script and the necessary files by invoking the tutorial command.
$ ssh username@crane.unl.edu
$ source osg_oasis_init
$ tutorial dagman-namd
$ cd tutorial-dagman-namd
The directory tutorial-dagman-namd contains all the necessary files. The file
linear.dag is the DAGMan script. The files namd_run_job0.submit, ... are the
HTCondor script files that execute the files namd_run_job0.sh,....
Let us take a look at the DAG file linear.dag.
$ nano linear.dag
######DAG file###### #comment
Job A0 namd_run_job0.submit #Job keyword, Job Name, Condor Job submission script.
Job A1 namd_run_job1.submit #Job keyword, Job Name, Condor Job submission script.
Job A2 namd_run_job2.submit #Job keyword, Job Name, Condor Job submission script.
Job A3 namd_run_job3.submit #Job keyword, Job Name, Condor Job submission script.
PARENT A0 CHILD A1 #Inter Dependency between Job A0 and A1
PARENT A1 CHILD A2 #Inter Dependency between Job A1 and A2
PARENT A2 CHILD A3 #Inter Dependency between Job A2 and A3
The first four lines after the comment are the listing of the condor jobs
with name assignment: A0, A1, A2 and A3. Here the condor job submit files are
namd_run_job0.submit, namd_run_job1.submit... that run the individual
MD simulations. The next three lines describe the relationship
among the four jobs.
Now we submit the DAGMan job.
$ condor_submit_dag linear.dag
-----------------------------------------------------------------------
File for submitting this DAG to Condor : linear.dag.condor.sub
Log of DAGMan debugging messages : linear.dag.dagman.out
Log of Condor library output : linear.dag.lib.out
Log of Condor library error messages : linear.dag.lib.err
Log of the life of condor_dagman itself : linear.dag.dagman.log
Submitting job(s).
1 job(s) submitted to cluster 1317501.
-----------------------------------------------------------------------
Note that the DAG file is submitted through condor_submit_dag.
Let's monitor the job status every two seconds. (Recall connect watch
from a previous lesson.)
$ watch -n 1 condor_q -dag username
-- Submitter: login.crane.hcc.unl.edu : <129.93.227.113:9619?noUDP&sock=205992_4558_3> : login.crane.hcc.unl.edu
ID OWNER/NODENAME SUBMITTED RUN_TIME ST PRI SIZE CMD
126476.0 username 1/7 10:01 0+00:00:20 R 0 0.3 condor_dagman -f -
126477.0 |-A0 1/7 10:01 0+00:00:00 I 0 0.0 namd_run_job0.sh
2 jobs; 0 completed, 0 removed, 1 idle, 1 running, 0 held, 0 suspended
We need to type Ctrl-C to exit from watch command. We see two running jobs. One is the dagman
job which manages the execution of NAMD jobs. The other is the actual NAMD
execution namd_run_job0.sh. Once the dag completes, you will see four .tar.gz
files OutFilesFromNAMD_job0.tar.gz, OutFilesFromNAMD_job1.tar.gz, OutFilesFromNAMD_job2.tar.gz,
OutFilesFromNAMD_job3.tar.gz. If the output files are not empty, the jobs are
successfully completed. Of course, a through check up requires looking at the output results.
Just one thing to note here, we used the -dag option with condor_q. This is a special option that tells HTCondor to
do a bit more processing and to present jobs grouped by the DAG that is running them. It doesn't help much in our
linear DAG example but with more complex DAGs, it makes it easier to see what jobs are associated with
a given DAG and how much of the DAG has been completed.
Parallel DAG
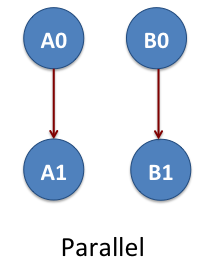
Now we consider the workflow of two-linear set of jobs A0, A1, B0 and B1. Again these are NAMD jobs. The job A0 is parent of A0 and the job B0 is the parent of B1. The jobs A0 and A1 do not depend on B0 and B1. This means we have two parallel DAGs that are represented as A0->A1 and B0->B1. The arrow shows the data dependency between the jobs. This example is located at
$ cd tutorial-dagman-namd/TwoLinearDAG
The directory contains the input files, job submission files and execution scripts of the
jobs. What is missing here is the .dag file. See if you can write the DAGfile for this example
and submit the job.
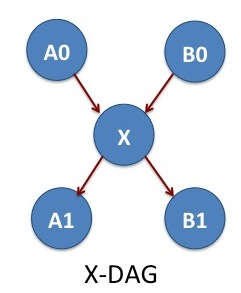
X-DAG
We consider one more example of jobs A0, A1, X, B0 and B1 that allows the cross communication between two parallel jobs. The jobs A0 and B0 are two-independent NAMD simulations. After finishing A0 and B0, we do some analysis with the job X. The jobs A1 and B1 are two MD simulations independent of each other. The job X determines what is the simulation temperature of MD simulations A1 and B1. In the DAGMan language, X is the parent of A1 and B1.
The input files, job submission files and execution scripts of the jobs are located at
$ cd tutorial-dagman-namd/X-DAG
Again we are missing the .dag file here. See if you can write the DAGfile for
this example
Job Retry and Rescue
In the above examples, the set of jobs have simple inter relationship. Indeed, DAGMan is capable of dealing with set of jobs with complex inter relations. One may also write a DAG file for set of DAG files where each of the DAG file contains the workflow for set of condor jobs. Also DAGMan can help with the resubmission of uncompleted portions of a DAG, when one or more nodes result in failure.
Job Retry
Say for example, job A2 in the linear dag example is important and you want to make sure that it completes. One way is to re-try the specific job A2 a few times. DAGMan would re-try failed jobs when you specify the following line at the end of dag file.
$ nano linear.dag #open the linear.dag file
### At the end of the linear.dag file
Retry A2 3 #This means re-try job A2 for three times in case of failures.
# If you want to retry jobs A2 and A3 for 7 times, edit the linear.dag
### At the end of the linear.dag file
Retry A2 7 #This means re-try job A2 for seven times in case of failures.
Retry A3 7 #This means re-try job A3 for seven times in case of failures.
Rescue DAG
In case DAGMan can not complete the set of jobs, it would create a rescue DAG
file with a suffix .rescue. The rescue DAG file contains the information about
where to restart the jobs. Say for example, in our workflow of four linear jobs,
the jobs A0 and A1 are finished and A2 is incomplete. In such a case we do not
want to start executing the jobs all over again rather we want to start from Job
A2. This information is embedded in the rescue dag file. In our example of
linear.dag, the rescue dag file would be linear.dag.rescue. So we re-submit
the rescue dag file as follows
$ condor_submit_dag linear.dag.rescue
Keypoints
- DAGMan (Directed Acyclic Graph Manager) handles computational jobs that are mapped as a directed acyclic graph.
- condor_submit_dag - is the command to submit a dagman job.
- One may write a DAGMan file consisting of several DAGMan jobs.
