Job Scheduling with HTCondor
Overview
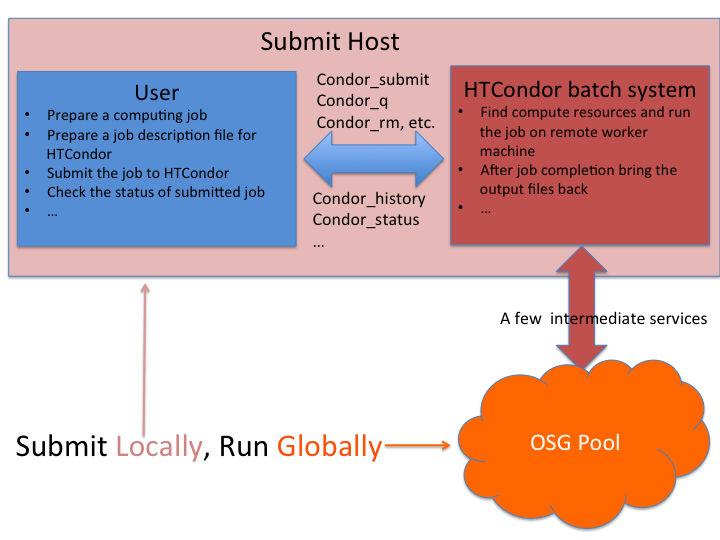
In this section, we will learn the basics of HTCondor for submitting and monitoring jobs. The jobs are submitted through the OSG Connect login node. The submitted jobs are executed on the remote worker node(s) and the outputs are transfered back to the login node.
Login to OSG Connect
using SSH:
# username is your CUE username
$ ssh username@scosg16.jlab.org
# enter your password
password:
You may receive a warning such as:
The authenticity of host 'scosg16.jlab.org (192.170.227.119)' can't be established.
RSA key fingerprint is SHA256:KRH0+kF1V5kNookplCt2f+lH4dKaZLowKbEevNnVmKY.
Are you sure you want to continue connecting (yes/no)?
This is normal when connecting to a new host for the first time. If prompted, type yes.
Copy tutorial files
Use the command below to copy the tutorial files into your account:
# creates a directory "OSG-UserTraining-JLab-2019"
$ git clone https://github.com/swc-osg-workshop/OSG-UserTraining-JLab-2019/
# enter the "OSG-UserTraining-JLab-2019" directory
$ cd OSG-UserTraining-JLab-2019
# entering the "tutorial-quickstart" directory
$ cd tutorial-quickstart
Job 1: A simple, nonparallel job
Inside the tutorial directory that you created or installed previously,
let's create a test script to execute as your job. For pretyped setup, this is
the short.sh file:
$ cat short.sh
#!/bin/bash
# short.sh: a short discovery job
printf "Start time: "; /bin/date
printf "Job is running on node: "; /bin/hostname
printf "Job running as user: "; /usr/bin/id
printf "Job is running in directory: "; /bin/pwd
echo
echo "Working hard..."
sleep 20
echo "Science complete!"
Now, make the script executable.
chmod +x short.sh
Run the job locally
When setting up a new job submission, it's important to test your job outside of HTCondor before submitting into the grid.
$ ./short.sh
Start time: Wed Aug 21 09:21:35 CDT 2013
Job is running on node: login01.osgconnect.net
Job running as user: uid=54161(netid) gid=1000(users) groups=1000(users),0(root),1001(osg-connect),1002(osg-staff),1003(osg-connect-test),9948(staff),19012(osgconnect)
Job is running in directory: /home/netid/quickstart
Working hard...
Science complete!
Create an HTCondor submit file
So far, so good! Let's create a simple (if verbose) HTCondor submit file. This can be found in tutorial01.submit.
$ cat tutorial01.submit
# Our executable is the main program or script that we've created
# to do the 'work' of a single job.
executable = short.sh
# We need to name the files that HTCondor should create to save the
# terminal output (stdout) and error (stderr) created by our job.
# Similarly, we need to name the log file where HTCondor will save
# information about job execution steps.
error = short.error
output = short.output
log = short.log
# We need to request the resources that this job will need:
request_cpus = 1
request_memory = 1 MB
request_disk = 1 MB
# The last line of a submit file indicates how many jobs of the above
# description should be queued. We'll start with one job.
queue 1
Submit the job
Submit the job using condor_submit:
$ condor_submit tutorial01.submit
Submitting job(s).
1 job(s) submitted to cluster 144121.
Check the job status
The condor_q command tells the status of currently running jobs. By default,
it should just show your jobs, although you can also use your username to show
only your jobs.
$ condor_q username
-- Schedd: login03.osgconnect.net : <192.170.227.22:9618?... @ 12/10/18 14:19:08
OWNER BATCH_NAME SUBMITTED DONE RUN IDLE TOTAL JOB_IDS
username ID: 1441271 12/10 14:18 _ 1 _ 1 1441271.0
1 jobs; 0 completed, 0 removed, 0 idle, 1 running, 0 held, 0 suspended
You can also get status on a specific job cluster:
$ condor_q 1441271
-- Schedd: login03.osgconnect.net : <192.170.227.22:9618?... @ 12/10/18 14:19:08
OWNER BATCH_NAME SUBMITTED DONE RUN IDLE TOTAL JOB_IDS
username ID: 1441271 12/10 14:18 _ 1 _ 1 1441271.0
1 jobs; 0 completed, 0 removed, 0 idle, 1 running, 0 held, 0 suspended
Note the DONE, RUN, and IDLE columns. Your job will be listed in the IDLE column if
it hasn't started yet. If it's currently scheduled and running, it will
appear in the RUN column. As it finishes up, it will then show in the DONE column.
Once the job completes completely, it will not appear in condor_q.
Job history
Once your job has finished, you can get information about its execution
from the condor_history command:
$ condor_history 1441272
ID OWNER SUBMITTED RUN_TIME ST COMPLETED CMD
1441272.0 netid 12/10 14:18 0+00:00:29 C 12/10 14:19 /home/netid/tutorial-quickstart/short.sh
Note: You can see much more information about your job's final status
using the -long option.
Check the job output
Once your job has finished, you can look at the files that HTCondor has returned to the working directory. The names of these files were specified in our submit file. If everything was successful, it should have returned:
- a log file from HTCondor for the job cluster: short.log
- an output file for each job's output: short.output
- an error file for each job's errors: short.error
Read the output file. It should be something like this:
$ cat short.output
Start time: Mon Dec 10 20:18:56 UTC 2018
Job is running on node: osg-84086-0-cmswn2030.fnal.gov
Job running as user: uid=12740(osg) gid=9652(osg) groups=9652(osg)
Job is running in directory: /srv
Working hard...
Science complete!
Job 2: Passing arguments to executables
Sometimes it's useful to pass arguments to your executable from your submit file. For example, you might want to use the same job script for more than one run, varying only the parameters. You can do that by adding arguments to your submission file.
We have modified our existing short.sh script to accept arguments. We can
see these changes in the short_transfer.sh
$ cat short_transfer.sh
#!/bin/bash
# short.sh: a short discovery job
printf "Start time: "; /bin/date
printf "Job is running on node: "; /bin/hostname
printf "Job running as user: "; /usr/bin/id
printf "Job is running in directory: "; /bin/pwd
printf "The command line argument is: "; $1
printf "Contents of $1 is "; cat $1
cat $1 > output.txt
printf "Working hard..."
ls -l $PWD
sleep 20
echo "Science complete!"
We need to make our new script executable just as we did before:
$ chmod +x short_transfer.sh
Notice that with our changes, the new script will now print out the contents of whatever file we specify in our arguments, specified by the $1. It will also copy the contents of that file into another file called output.txt.
We have a simple text file called input.txt that we can pass to our script:
$ cat input.txt
"Hello World"
Once again, before submitting our job we should test it locally to ensure it runs as we expect:
$ ./short_transfer.sh input.txt
Start time: Tue Dec 11 10:19:12 CST 2018
Job is running on node: login03.osgconnect.net
Job running as user: uid=100279(netid) gid=1000(users) groups=1000(users),5532(connect),5782(osg),7021(osg.ConnectTrain)
Job is running in directory: /home/netid/tutorial-quickstart
The command line argument is: Contents of input.txt is "Hello World"Working hard...total 28
drwxrwxr-x 2 netid users 34 Oct 15 09:37 Images
-rw-rw-r-- 1 netid users 13 Oct 15 09:37 input.txt
drwxrwxr-x 2 netid users 114 Dec 11 09:50 log
-rw-r--r-- 1 netid users 13 Dec 11 10:19 output.txt
-rwxrwxr-x 1 netid users 291 Oct 15 09:37 short.sh
-rwxrwxr-x 1 netid users 390 Dec 11 10:18 short_transfer.sh
-rw-rw-r-- 1 netid users 806 Oct 15 09:37 tutorial01.submit
-rw-rw-r-- 1 netid users 547 Dec 11 09:49 tutorial02.submit
-rw-rw-r-- 1 netid users 1321 Oct 15 09:37 tutorial03.submit
Science complete!
Now, we need to edit our submit file to properly handle these new arguments and output files.
This is saved as tutorial02.submit
$ cat tutorial02.submit
# We need the job to run our executable script, with the
# input.txt filename as an argument, and to transfer the
# relevant input and output files:
executable = short_transfer.sh
arguments = input.txt
transfer_input_files = input.txt
transfer_output_files = output.txt
error = job.error
output = job.output
log = job.log
# The below are good base requirements for first testing jobs on OSG,
# if you don't have a good idea of memory and disk usage.
request_cpus = 1
request_memory = 1 GB
request_disk = 1 GB
# Queue one job with the above specifications.
queue 1
Notice the added arguments = input.txt information. The arguments option specifies what arguments should be passed to the executable.
The transfer_input_files and transfer_output_files options need to be included as well. When jobs are deployed on the Open Science Grid, they are sent only with files that are specified. Additionally, only the specified output files are returned with the job. Any output not transferred back, with the exception of our error, output, and log files, are discarded at the end of the job.
Submit the new submit file using condor_submit. Be sure to check your output files once the job completes.
$ condor_submit tutorial02.submit
Submitting job(s).
1 job(s) submitted to cluster 1444781.
Job 3: Submitting jobs concurrently
What do we need to do to submit several jobs simultaneously? In the
first example, Condor returned three files: out, error, and log. If we
want to submit several jobs, we need to track these three files for each
job. An easy way to do this is to add the $(Cluster) and $(Process)
macros to the HTCondor submit file. Since this can make our working
directory really messy with a large number of jobs, let's tell HTCondor
to put the files in a directory called log. Here's what the third submit file looks like, called tutorial03.submit:
$ cat tutorial03.submit
# We need the job to run our executable script, arguments and files.
# Also, we'll specify unique filenames for each job by using
# the job's 'cluster' value.
executable = short_transfer.sh
arguments = input.txt
transfer_input_files = input.txt
transfer_output_files = output.txt
error = log/job.$(Cluster).$(Process)error
output = log/job.$(Cluster).$(Process).output
log = log/job.$(Cluster).$(Process).log
request_cpus = 1
request_memory = 1 GB
request_disk = 1 GB
# Let's queue ten jobs with the above specifications
queue 10
Before submitting, we also need to make sure the log directory exists.
$ mkdir -p log
You'll see something like the following upon submission:
$ condor_submit tutorial03.submit
Submitting job(s)..........
10 job(s) submitted to cluster 1444786.
Look at the output files in the log directory and notice how each job received its own separate output file:
$ ls ./log
job.1444786.0.error job.1444786.1.error job.1444786.2.error job.1444786.3.error job.1444786.4.error job.1444786.5.error job.1444786.6.error job.1444786.7.error job.1444786.8.error job.1444786.9.error
job.1444786.0.log job.1444786.1.log job.1444786.2.log job.1444786.3.log job.1444786.4.log job.1444786.5.log job.1444786.6.log job.1444786.7.log job.1444786.8.log job.1444786.9.log
job.1444786.0.output job.1444786.1.output job.1444786.2.output job.1444786.3.output job.1444786.4.output job.1444786.5.output job.1444786.6.output job.1444786.7.output job.1444786.8.output job.1444786.9.output
Where did jobs run?
When we start submitting a lot of simultaneous jobs into the queue, it might
be worth looking at where they run. To get that information, we'll use a
couple of condor_history commands. First, run condor_history -long jobid
for your first job. Again the output is quite long:
$ condor_history -long 1444786
BlockWriteKbytes = 0
BlockReads = 0
DiskUsage_RAW = 36
...
Looking through here for a hostname, we can see that the parameter
that we want to know is LastRemoteHost. That's what job slot our job
ran on. With that detail, we can construct a shell command to get
the execution node for each of our 100 jobs, and we can plot the
spread. LastRemoteHost normally combines a slot name and a host name,
separated by an @ symbol, so we'll use the UNIX cut command to slice off
the slot name and look only at hostnames. We'll cut again on the period
in the hostname to grab the domain where the job ran.
For illustration, the author has submitted a thousand jobs for a more interesting distribution output.
$ condor_history -format '%s\n' LastRemoteHost 942 | cut -d@ -f2 | distribution --height=100
Val |Ct (Pct) Histogram
[netid@login01 log]$ condor_history -format '%s\n' LastRemoteHost 959 | cut -d@ -f2 | cut -d. -f2,3 | distribution --height=100
Val |Ct (Pct) Histogram
mwt2.org |456 (46.77%) +++++++++++++++++++++++++++++++++++++++++++++++++++++
uchicago.edu |422 (43.28%) +++++++++++++++++++++++++++++++++++++++++++++++++
local |28 (2.87%) ++++
t2.ucsd |23 (2.36%) +++
phys.uconn |12 (1.23%) ++
tusker.hcc |10 (1.03%) ++
...
The distribution program reduces a list of hostnames to a set of
hostnames with no duplication (much like sort | uniq -c), but
additionally plots a distribution histogram on your terminal
window. This is nice for seeing how Condor selected your execution
endpoints.
There is also condor_plot a command that plots similar information in a
HTML page. You can have bar plots, pie charts and more.
Removing jobs
On occasion, jobs will need to be removed for a variety of reasons
(incorrect parameters, errors in submission, etc.). In these instances,
the condor_rm command can be used to remove an entire job submission
or just particular jobs in a submission. The condor_rm command accepts
a cluster id, a job id, or username and will remove an entire cluster
of jobs, a single job, or all the jobs belonging to a given user
respectively. E.g. if a job submission generates 100 jobs and is
assigned a cluster id of 103, then condor_rm 103.0 will remove the
first job in the cluster. Likewise, condor_rm 103 will remove all
the jobs in the job submission and condor_rm [username] will remove
all jobs belonging to the user. The condor_rm documenation has more
details on using condor_rm including ways to remove jobs based on other
constraints.
