Introduction to the Open Science Grid
The Open Science Grid
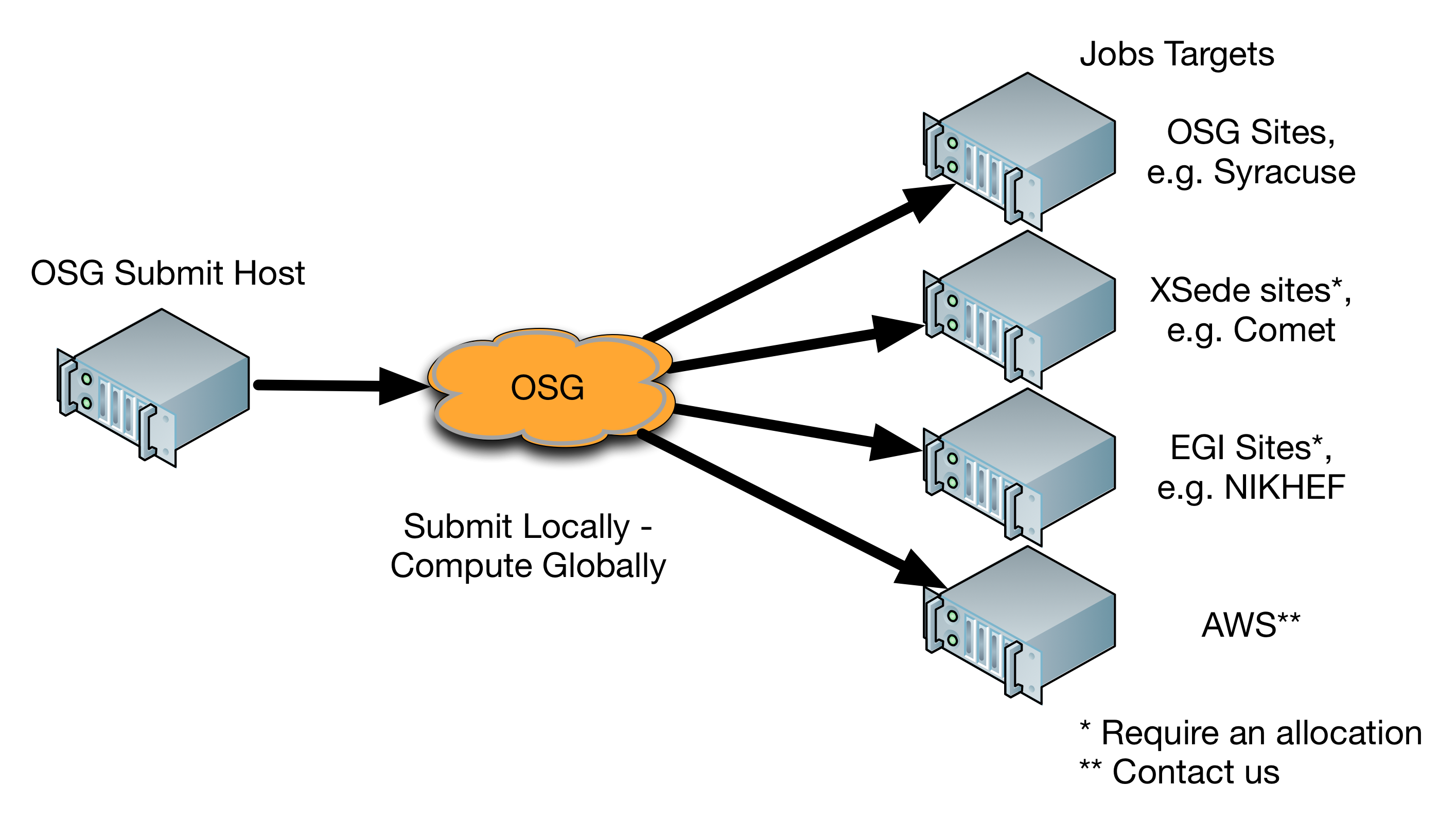
The Open Science Grid (OSG) is a consortium of research communities that provides resources to use distributed high throughput computing (HTC) for scientific research. The OSG:
- Enables distributed computing on backfill capacity at more than 120 institutions.
- Delivers nearly 2 million core CPU hours per day, including ~500,000 'opportunistic' hours available to campuses and individual research groups (though this varies significantly from day-to-day, and there's even more un-tapped capacity available).
- Allows campuses to join the consortium by (1) contributing their own backfill capacity (from just about any 'type' of Linux cluster), and/or (2) establishing a local submission point for local researchers to access OSG.
- Provides the OSG Connect service for individual research groups who do not have a local OSG submission point.
- Supports a variety of common research applications in a core OSG-maintained repository (called OASIS).
- Supports several options for working with large, per-job input and output.
- Leverages the HTC-optimized HTCondor compute scheduling software for job queueing and execution (on top of a layer of OSG-specific softwae and tools).
Computation that is a good match for OSG
Computational workloads that are accelerated with an HTC execution approach are those that can parallelize naturally to numerous, completely independent 'jobs', and this turns out to be the case for VERY many research workloads, especially those typically defined as 'big data'. For example, parameter space optimizations, image processing, various forms of text processing (including much of computational genomics and bioinformatics), and sets of numerous, relatively-small simulations can all be executed as numerous, independent jobs whose results can be later combined or analyzed. Nearly every time a researcher has written their own code with an internal 'mapping' or 'loop' step that is the very time-intensive, they will see far greater throughput by using an HTC execution approach and be able to expand the 'space' of their computational work to tackle greater research challenges.
The best HTC workloads for OSG can be executed as many jobs that EACH complete on 1 CPU core in less than 12 hours, with less than 100 MB of input or output data. The HTCondor queueing system can transfer all executables, input data, and output data (up to a certain size) from the submit server to the job, as specified in job submit files created by the user. However, the technologies in OSG also support jobs needing multiple cores, GPUs, and larger per-job data. Generally, the 'less' each job needs, the more jobs a user will have running, and sooner. Jobs requesting 1 CPU core and less than 1 GB of memory will typically get hundreds or up to thousands of cores running their jobs in the OSG, and reach that maximum within several hours of initially submitting the jobs.
Another component of 'fit' for jobs running on OSG pertains to the necessary software and computing environment. Jobs submitted to the OSG will be executed at several remote clusters (OSG is a distributed HTC, or 'dHTC' environment). The execute servers at each of these will differ a bit from the computing environment on the submit server, but all of OSG execute servers currently run RedHat-compatible Linux of major version 6 (decreasingly) or 7. This heterogeneity means that jobs will run most consistently if they bring along the major portions of the software environment they'll need (or leverage OSG-provided software for common applications). Jobs that leverage statically-compiled binaries (which includes many genomics applications, for example) are the most robust across the environment heterogeneity in OSG, but there are many users whose jobs bring along a pre-compiled version of Python or R, or leverage Matlab's runtime interpreter for compiled Matlab code, as just a few examples.
Consider the following dHTC guidelines:
- Jobs should run single-threaded, individually using less than 2 GB memory and running for 1-12 hours. There is support for jobs with longer run time, more memory or multi-threading support (just fewer 'slots' regularly becoming available for them, and jobs can be evicted at any time).
- Only core utilities can be expected on the remote end. There is no standard version of software even for
gcc,python,BLAS, or others. Consider using OSG-supported Software Modules, see below, to manage software dependencies, look through our High Throughput Computing Recipes, or get in touch with the OSG Connect team to discuss the best approach for 'packing up' your software dependencies. - Input and output data for each job should be < 10 GB to allow them to be transferred in by the jobs, processed and returned to the submit node. The scheduler, HTCondor, can transfer input and output files up to 100 MB (~500 MB total per job), and other transfer methods (
scp,rsync,GridFTP) are better suited for larger per-job data transfers. Please contact the user support listed below for more information
Computation that is NOT a good match for OSG
The following are examples of computations that are NOT good matches for OSG:
- Tightly coupled computations, for example using MPI-based communication across multiple compute nodes (due to the distributed nature of the OSG infrastructure).
- Computations requiring a shared file system, as there is no shared file system between the different clusters on the OSG.
- Computations requiring complex software deployments or proprietary software are not a good fit. There is limited support for distributing software to the compute clusters, and for complex software, or licensed software, deployment can be a nearly impossible task.
How to get help using or joining OSG
Please contact user support staff at user-support@opensciencegrid.org.
Available Resources on OSG
Commonly used open source software and libraries are available in a central repository (known as OASIS) and accessed via the module command on OSG. We will see how to search for, load, and use software modules. This command may be familiar to you if you have used HPC clusters before such as XSede's Comet or NERSC's Cori.
We will also cover the usage of the built-in tutorial command. Using tutorial, we load a variety of job templates that cover basic usage, specific use cases, and best practices.
Software Modules
To take a look at the module command, log in to the OSG Connect submit host via SSH:
$ ssh username@training.osgconnect.net
Once you are logged in, you can check the available modules:
$ module avail
[...]
ANTS/1.9.4 dmtcp/2.5.0 lapack/3.5.0 protobuf/2.5
ANTS/2.1.0 (D) ectools lapack/3.6.1 (D) psi4/0.3.74
MUMmer3.23/3.23 eemt/0.1 libXpm/3.5.10 python/2.7 (D)
OpenBUGS/3.2.3 elastix/2015 libgfortran/4.4.7 python/3.4
OpenBUGS-3.2.3/3.2.3 espresso/5.1 libtiff/4.0.4 python/3.5.2
R/3.1.1 (D) espresso/5.2 (D) llvm/3.6 qhull/2012.1
R/3.2.0 ete2/2.3.8 llvm/3.7 root/5.34-32-py34
R/3.2.1 expat/2.1.0 llvm/3.8.0 (D) root/5.34-32
R/3.2.2 ffmpeg/0.10.15 lmod/5.6.2 root/6.06-02-py34 (D)
R/3.3.1 ffmpeg/2.5.2 (D) madgraph/2.1.2 rosetta/2015
R/3.3.2 fftw/3.3.4-gromacs madgraph/2.2.2 (D) rosetta/2016-02
RAxML/8.2.9 fftw/3.3.4 (D) matlab/2013b rosetta/2016-32 (D)
SeqGen/1.3.3 fiji/2.0 matlab/2014a ruby/2.1
[...]
Where:
(D): Default Module
Use "module spider" to find all possible modules.
Use "module keyword [key1 key2...]" to search for all possible modules matching any of the "keys".
In order to load a module, you need to run module load [modulename]. If you want to load the R package,
$ module load R
This sets up the R package for you. Now you can do a test calculations with R.
# invoke R
$ R
# simple on-screen calculation with cosine function
> cos(45)
[1] 0.525322
> quit()
Save workspace image? [y/n/c]: n
If you want to unload a module, type
$ module unload R
For a more complete list of all available modules please check the support page or module spider. Local submit nodes and OSG member institutions can also leverage the same set of OSG-supported software modules.
Tutorial Command
The built-in tutorial command assists a user in getting started on OSG Connect servers. To see the list of existing tutorials, type
$ tutorial
Say, for example, you are interested in learning how to run R scripts on OSG, the
tutorial command sets up the R tutorial for you.
$ tutorial R
Installing R (master)...
Tutorial files installed in ./tutorial-R.
Running setup in ./tutorial-R...
The tutorial R command creates a directory tutorial-R containing the necessary script and input files.
# The example R script file
mcpi.R
# The job execution file
R-wrapper.sh
# The job submission file (will discuss later in the lesson HTCondor scripts)
R.submit
Let's focus on mcpi.R and the R wrapper scripts. The details of R.submit script will be discussed later when we learn about submitting jobs with HTCondor.
The file mcpi.R is an R script that calculates the value of pi using the Monte Carlo method. The R-wrapper.sh essentially loads the R module and runs the mcpi.R
script.
#!/bin/bash
EXPECTED_ARGS=1
if [ $# -ne $EXPECTED_ARGS ]; then
echo "Usage: R-wrapper.sh file.R"
exit 1
else
source /cvmfs/oasis.opensciencegrid.org/osg/modules/lmod/current/init/bash
module load R
Rscript $1
fi
There are other tutorials available, which can serve as templates to develop your own scripts to run your workloads on OSG.
